Bayesian Linear Model
Caleb Jin / 2019-04-18
Linear model
Consider a multiple linear regression model as follows: y=Xβ+ϵ,y=Xβ+ϵ, where y=(y1,y2,…,yn)Ty=(y1,y2,…,yn)T is the nn-dimensional response vector, X=(x1,x2,…,xn)TX=(x1,x2,…,xn)T is the n×pn×p design matrix, and ϵ∼Nn(0,σ2In)ϵ∼Nn(0,σ2In). We assume that p<np<n and XX is full rank.
Maximum likelihood estimation (MLE) approach
Since y∼Nn(Xβ,σ2In)y∼Nn(Xβ,σ2In), the likelihood function is given as L(β,σ2)=f(y|β,σ2)=(2π)−n2|Σ|−12exp{−12(y−Xβ)TΣ−1(y−Xβ)},L(β,σ2)=f(y|β,σ2)=(2π)−n2|Σ|−12exp{−12(y−Xβ)TΣ−1(y−Xβ)},
where Σ=σ2InΣ=σ2In. Then the log likelihood can be written as l(β,σ2)=logL(β,σ2)=−n2log(2π)−n2log(σ2)−12σ2(y−Xβ)T(y−Xβ).l(β,σ2)=logL(β,σ2)=−n2log(2π)−n2log(σ2)−12σ2(y−Xβ)T(y−Xβ). Note that l(β,σ2)l(β,σ2) is a concave function in (β,σ2)(β,σ2). Hence, the maximum likelihood estimator (MLE) can be obtained by solving the following equations: ∂l(β,σ2)∂β=−12σ2(−2XTy+2XTXβ)=0;∂l(β,σ2)∂σ2=−n21σ2+121(σ2)2(y−Xβ)T(y−Xβ)=0.∂l(β,σ2)∂β=−12σ2(−2XTy+2XTXβ)=0;∂l(β,σ2)∂σ2=−n21σ2+121(σ2)2(y−Xβ)T(y−Xβ)=0. Therefore, the MLEs of ββ and σ2σ2 are given as ˆβ=(XTX)−1XTy;ˆσ2=(y−Xˆβ)T(y−Xˆβ)n=‖y−ˆy‖2n,^β=(XTX)−1XTy;^σ2=(y−X^β)T(y−X^β)n=∥y−^y∥2n, where ˆy=Xˆβ^y=X^β.
Distribution of ˆβ^β and ˆσ2^σ2
Note that if z∼Nk(μ,Σ)z∼Nk(μ,Σ), then Az∼Nm(Aμ,AΣAT)Az∼Nm(Aμ,AΣAT), where AA is an m×km×k matrix. Since y∼Nn(Xβ,σ2In)y∼Nn(Xβ,σ2In) and ˆβ=(XTX)−1XTy^β=(XTX)−1XTy, we have ˆβ∼Np((XTX)−1XTXβ,σ2(XTX)−1XTX(XTX)−1)=Np(β,σ2(XTX)−1).^β∼Np((XTX)−1XTXβ,σ2(XTX)−1XTX(XTX)−1)=Np(β,σ2(XTX)−1).
Note that y−ˆy=(In−X(XTX)−1XT)yy−^y=(In−X(XTX)−1XT)y, where In−X(XTX)−1XTIn−X(XTX)−1XT is an idempotent matrix with rank (n−p)(n−p).
Can you prove that In−X(XTX)−1XT s an idempotent matrix of rank (n−p) ?Can you prove that In−X(XTX)−1XT s an idempotent matrix of rank (n−p) ?
Proof. Let H=X(XTX)−1XTH=X(XTX)−1XT.
HH=X(XTX)−1XTX(XTX)−1XT=X(XTX)−1XT=HHH=X(XTX)−1XTX(XTX)−1XT=X(XTX)−1XT=H thus HH is idempotent matrix.
Similarly, as (In−H)(In−H)=In−H(In−H)(In−H)=In−H, (In−H)(In−H) is also idempotent.
Hence we have r(In−H)=tr(In−H)=n−tr(H)=n−tr((XTX)−1XTX)=n−pr(In−H)=tr(In−H)=n−tr(H)=n−tr((XTX)−1XTX)=n−pHow to prove r(In−H)=tr(In−H)? How to prove r(In−H)=tr(In−H)?
The eigenvalue of idempotent matrix is either 1 or 0, hence the rank of it is the sum of eigenvalues, which equals the trace of matrix;
How to prove trace of matrix is the sum of eigenvalues?How to prove trace of matrix is the sum of eigenvalues?
It requires characteristic polynomial.
Another way to prove this is in this link
From Lemma 1, we have that nˆσ2σ2∼χ2(n−p),n^σ2σ2∼χ2(n−p),(1) where χ2(n−p)χ2(n−p) denotes the chi-squared distribution with degrees of freedom n−pn−p.
Can you prove Eq.Can you prove Eq.(1)?
Proof. By Lemma 1, we have
nˆσ2σ2=ˆeTˆeσ2=yT(In−Hσ2)y=yTAy∼χ2(tr(AΣ),μTAμ/2)n^σ2σ2=^eT^eσ2=yT(In−Hσ2)y=yTAy∼χ2(tr(AΣ),μTAμ/2)
Bayesian approach
σ2σ2 is known
Suppose σ2σ2 is known. We define the prior distribution of ββ by β∼Np(0,σ2νIp)β∼Np(0,σ2νIp). Then the posterior density of ββ can be obtained by π(β|y)∝f(y|β)π(β)∝exp(−12σ2‖y−Xβ‖2)×exp(−12σ2ν‖β|2)=exp[−12σ2{(y−Xβ)T(y−Xβ)+1νβTβ}]∝exp{−12σ2(−2βTXTy+βTXTXβ+1νβTβ)}=exp{−12σ2(−2βT(XTX+1νIp)(XTX+1νIp)−1XTy+βT(XTX+1νIp)β)}∝exp{−12σ2(β−˜β)T(XTX+1νIp)(β−˜β)}π(β|y)∝f(y|β)π(β)∝exp(−12σ2∥y−Xβ∥2)×exp(−12σ2ν∥β|2)=exp[−12σ2{(y−Xβ)T(y−Xβ)+1νβTβ}]∝exp{−12σ2(−2βTXTy+βTXTXβ+1νβTβ)}=exp{−12σ2(−2βT(XTX+1νIp)(XTX+1νIp)−1XTy+βT(XTX+1νIp)β)}∝exp{−12σ2(β−~β)T(XTX+1νIp)(β−~β)} where ˜β=(XTX+1ν)−1XTy~β=(XTX+1ν)−1XTy.
This implies that β|y∼Np((XTX+1νIp)−1XTy,σ2(XTX+1νIp)−1).β|y∼Np((XTX+1νIp)−1XTy,σ2(XTX+1νIp)−1).(2) The Bayesian estimate ˆβBayes=(XTX+1νIp)−1XTy^βBayes=(XTX+1νIp)−1XTy. It is worth noting that if ν→∞ν→∞, the posterior distribution converges to the distribution of ˆβMLE∼Np(β,σ2(XTX)−1)^βMLE∼Np(β,σ2(XTX)−1).
σ2σ2 is unknown
In general, σ2σ2 is unknown. Then, we need to assign a reasonable prior distribution for σ2σ2. We consider the inverse gamma distribution, which is called a , as follows: σ2∼IG(a0,b0)σ2∼IG(a0,b0) with the density function π(σ2)=ba00Γ(a0)(σ2)−a0−1exp(−b0σ2),π(σ2)=ba00Γ(a0)(σ2)−a0−1exp(−b0σ2), where a0>0a0>0 and b0>0b0>0. In addition, we need to introduce prior for β|σ2β|σ2:
β|σ2∼Np(0,σ2νIp).β|σ2∼Np(0,σ2νIp).
Today, we derive the joint posterior distributionπ(β,σ2|y)∝f(y|β,σ)π(β|σ2)π(σ2).Today, we derive the joint posterior distributionπ(β,σ2|y)∝f(y|β,σ)π(β|σ2)π(σ2).
Show
- π(σ2|β,y)=IG(a∗,b∗)π(σ2|β,y)=IG(a∗,b∗)
- π(β|y)∼π(β|y)∼ t-distribution with t∗t∗
Proof (1). π(σ2|β,y)=f(y|β,σ2)π(β,σ2)∫f(y|β,σ2)π(β,σ2)dσ2=f(y|β,σ2)π(β|σ2)π(σ2)∫f(y,β,σ2)dσ2∝f(y|β,σ2)π(β|σ2)π(σ2)∝(σ2)−n2exp(−12σ2‖y−Xβ‖2)×(σ2)−p2exp(−12σ2ν‖β‖2)×(σ2)−a0−1exp(−b0σ2)=(σ2)−(n+p2+a0)−1exp[−1σ2{12‖y−Xβ‖2+12ν‖β‖2+b0}]=(σ2)−a∗−1exp(−b∗σ2)π(σ2|β,y)=f(y|β,σ2)π(β,σ2)∫f(y|β,σ2)π(β,σ2)dσ2=f(y|β,σ2)π(β|σ2)π(σ2)∫f(y,β,σ2)dσ2∝f(y|β,σ2)π(β|σ2)π(σ2)∝(σ2)−n2exp(−12σ2∥y−Xβ∥2)×(σ2)−p2exp(−12σ2ν∥β∥2)×(σ2)−a0−1exp(−b0σ2)=(σ2)−(n+p2+a0)−1exp[−1σ2{12∥y−Xβ∥2+12ν∥β∥2+b0}]=(σ2)−a∗−1exp(−b∗σ2) where a∗=n+p2+a0a∗=n+p2+a0, b∗=12‖y−Xβ‖2+12ν‖β‖2+b0b∗=12∥y−Xβ∥2+12ν∥β∥2+b0.
This implies that σ2|β,y∼IG(n+p2+a0,12‖y−Xβ‖2+12ν‖β‖2+b0).σ2|β,y∼IG(n+p2+a0,12∥y−Xβ∥2+12ν∥β∥2+b0).Proof (2). π(β|y)=∫π(β,σ2|y)dσ2=∫f(y|β,σ2)π(β,σ2)∬f(y|β,σ2)π(β,σ2)dβdσ2dσ2∝∫f(y|β,σ2)π(β|σ2)π(σ2)dσ2∝Γ(a∗)b−a∗∝b∗−a∗=[12‖y−Xβ‖2+12ν‖β‖2+b0]−a∗∝[(y−Xβ)T(y−Xβ)+1νβTβ−2b0]−a∗=[yTy−2βTXTy+βTXTXβ+1νβTβ+2b0]−a∗∝[βT(XTX+1νIp)β−2βT(XTX+1νIp)(XTX+1νIp)−1XTy+yTy+2b0]−a∗=[βTAβ−2βTA˜β+˜βTA˜β−˜βTA˜β+yTy+2b0]−a∗=[(β−˜β)TA(β−˜β)+yTy−yTXA−1XTy+2b0]−a∗=[(β−˜β)TA(β−˜β)+yT(In−XA−1XT)y+2b0]−a∗∝[(β−˜β)TA(β−˜β)+c∗]−a∗∝[1+1c∗(β−˜β)TA(β−˜β)]−n+p+2a02=[1+ν∗ν∗c∗(β−˜β)TA(β−˜β)]−p+ν∗2=[1+1ν∗(β−˜β)T(c∗ν∗A−1)−1(β−˜β)]−p+ν∗2π(β|y)=∫π(β,σ2|y)dσ2=∫f(y|β,σ2)π(β,σ2)∬f(y|β,σ2)π(β,σ2)dβdσ2dσ2∝∫f(y|β,σ2)π(β|σ2)π(σ2)dσ2∝Γ(a∗)b−a∗∝b∗−a∗=[12∥y−Xβ∥2+12ν∥β∥2+b0]−a∗∝[(y−Xβ)T(y−Xβ)+1νβTβ−2b0]−a∗=[yTy−2βTXTy+βTXTXβ+1νβTβ+2b0]−a∗∝[βT(XTX+1νIp)β−2βT(XTX+1νIp)(XTX+1νIp)−1XTy+yTy+2b0]−a∗=[βTAβ−2βTA~β+~βTA~β−~βTA~β+yTy+2b0]−a∗=[(β−~β)TA(β−~β)+yTy−yTXA−1XTy+2b0]−a∗=[(β−~β)TA(β−~β)+yT(In−XA−1XT)y+2b0]−a∗∝[(β−~β)TA(β−~β)+c∗]−a∗∝[1+1c∗(β−~β)TA(β−~β)]−n+p+2a02=[1+ν∗ν∗c∗(β−~β)TA(β−~β)]−p+ν∗2=[1+1ν∗(β−~β)T(c∗ν∗A−1)−1(β−~β)]−p+ν∗2 This implies that β|y∼MT(˜β,c∗ν∗A−1,ν∗),β|y∼MT(~β,c∗ν∗A−1,ν∗),(3)
where
A=XTX+1νIp,˜β=A−1XTy,c∗=yT(In−XA−1XT)y+2b0,ν∗=n+2a0.A=XTX+1νIp,~β=A−1XTy,c∗=yT(In−XA−1XT)y+2b0,ν∗=n+2a0.
Note: the density of a multiple t-distribution with Σ,μΣ,μ and νν is (3)
Γ[(ν+p)/2]Γ(ν/2)νp/2πp/2|Σ|1/2[1+1ν(X−μ)TΣ−1(X−μ)]−(ν+p)/2.Γ[(ν+p)/2]Γ(ν/2)νp/2πp/2|Σ|1/2[1+1ν(X−μ)TΣ−1(X−μ)]−(ν+p)/2.
Monte Carlo Simulation
Suppose σ2σ2 is unknown. If β∼Np(0,σ2νIp)β∼Np(0,σ2νIp), we know the distribution of β|yβ|y is Eq. (3). According to this known distribution, we can easily compute the E(β|y)E(β|y) and V(β|y)V(β|y). But in practice, the form of density function π(β|y)π(β|y) is often an unknown and very complicated distribution, leading to be impossible to solve its integration directly. hence we turn to use numerical methods to solve this problem, such as Monte Carlo methods.
For example, a form density function π(θ|y)π(θ|y) is an unknown distribution. E(θ|y)=∫θπ(θ|y)dθE(θ|y)=∫θπ(θ|y)dθ. According to lemma , ˉXn→μ=E(X)¯Xn→μ=E(X) as n→∞n→∞. Therefore, if we indepedently generate θ(1),θ(2),…,θ(M)θ(1),θ(2),…,θ(M) from π(θ|y)π(θ|y), M−1∑Mk=1θ(k)→E(θ|y)M−1∑Mk=1θ(k)→E(θ|y) as M →∞→∞. However, there are two problems.
What if we cannot generate sample from π(θ|y)π(θ|y)
What if they are not identically and independently distributed.
The solutions to these two question are Monte Carlo (MC) method and Markov Chain Monte Carlo (MCMC).
For the first question, we can use importance sampling method.Lemma 2 (Strong Law of Large Number)
Let X1,X2,...X1,X2,... be a sequence of i.i.d random variables, each having finite mean m. Then Pr(limn→∞1n(X1+X2+...+Xn=m))=1.Pr(limn→∞1n(X1+X2+...+Xn=m))=1.σ2σ2 is known
Importance Sampling
To estimate mean value of parameters, usually we randomly sample from π(θ|y)π(θ|y) and compute their mean value to estimate parameters. However, in practice, it is hard to generate sample directly from π(θ|y)π(θ|y), because we don’t know what speccific distribution it belongs to. hence we return to importance sampling method.
Note that E(θ|y)=∫θπ(θ|y)dθ=∫θπ(θ|y)g(θ)g(θ)dθ=∫h(θ)g(θ)dθ=E{h(θ)}E(θ|y)=∫θπ(θ|y)dθ=∫θπ(θ|y)g(θ)g(θ)dθ=∫h(θ)g(θ)dθ=E{h(θ)} where h(θ)=θπ(θ|y)g(θ),g(θ)h(θ)=θπ(θ|y)g(θ),g(θ) is a known pdf.
To estimate the variance of parameters, similarly, Var(θ|y)=E[θ−E(θ|y)]2=∫(θ−E(θ|y))2π(θ|y)dθ=∫(θ−E(θ|y))2π(θ|y)g(θ)g(θ)dθ=∫h′(θ)g(θ)dθ=E{h′(θ)}Var(θ|y)=E[θ−E(θ|y)]2=∫(θ−E(θ|y))2π(θ|y)dθ=∫(θ−E(θ|y))2π(θ|y)g(θ)g(θ)dθ=∫h′(θ)g(θ)dθ=E{h′(θ)} where h′(θ)=(θ−E(θ|y))2π(θ|y)g(θ),h′(θ)=(θ−E(θ|y))2π(θ|y)g(θ), and g(θ)g(θ) is a known pdf.
The importance sampling method can be implemented as follows:Note that M∑i=1h(θi)M→E(h(θ))=E(θ|y).asM→∞M∑i=1h(θi)M→E(h(θ))=E(θ|y).asM→∞ Estimating variance by importance sampling method is similarly.
Importance Sampling Simulation
Setup
Consider a single linear regression model as follows:
yi=β0+β1xi+εiyi=β0+β1xi+εi where εi∼N(0,1),xi∼N(0,1),β0=1,β1=2,εi∼N(0,1),xi∼N(0,1),β0=1,β1=2, for i=1,…,100.i=1,…,100.
We already know that if β∼Np(0,σ2νIp)β∼Np(0,σ2νIp), then the distribution of β|yβ|y is Eq.(2). Assume σ=1σ=1 is known and consider a known pdf π(β)=ϕ(β;(0,0),10I2)π(β)=ϕ(β;(0,0),10I2), hence in this case, ν=10ν=10. Our simulation can be broken down into 3 steps in following:
Find exact value of E(β0|y),E(β1|y),V(β0|y)E(β0|y),E(β1|y),V(β0|y) and V(β1|y)V(β1|y)
Use the MC method to simulate the results above with size of 100, 1000 and 5000.
Use Importance Sampling Method to simulate relevant results with the same size in (2). Consider the known pdf of parameters are ϕ(β0|1,0.5)ϕ(β0|1,0.5) and ϕ(β1|2,0.5).ϕ(β1|2,0.5).
Simulation Results
We use R softeware to do this simulation.
- According to Eq. (2), compute E(β|y)=(XTX+1νIp)−1XTyE(β|y)=(XTX+1νIp)−1XTy and Var(β|y)=σ2(XTX+1νIp)−1Var(β|y)=σ2(XTX+1νIp)−1,
where σ=1σ=1 and ν=10ν=10, then we get the following results:
E(β0)E(β0) E(β1)E(β1) Var(β0)Var(β0) Var(β1)Var(β1) 1.108 1.997 0.01 0.011 - Sample directly from normal distribution of Eq. (2) with sample size of 100, 1000 and 5000, then compute the mean and variance values of samples. hence we get the following results:
| E(β0)E(β0) | E(β1)E(β1) | Var(β0)Var(β0) | Var(β1)Var(β1) | |
|---|---|---|---|---|
| 100 | 1.104 | 2.002 | 0.009 | 0.012 |
| 1000 | 1.107 | 1.993 | 0.011 | 0.012 |
| 5000 | 1.108 | 1.996 | 0.010 | 0.011 |
- Sample β0β0 and β1β1 directly from ϕ(β0|1,0.5)ϕ(β0|1,0.5) and ϕ(β1|2,0.5)ϕ(β1|2,0.5), respectively, with sample size of 100, 1000 and 5000. And then compute h(θi)h(θi) and
h′(θi)h′(θi). Compute mean values of them to get estimates of expectation and variance. The
final results are following:
E(β0)E(β0) E(β1)E(β1) Var(β0)Var(β0) Var(β1)Var(β1) 100 1.0500 2.0623 0.0175 0.0127 1000 1.1323 1.9521 0.0106 0.0127 5000 1.1269 2.0478 0.0107 0.0137
σ2σ2 is unknown
Suppose σ2σ2 is unknown, we cannot use π(β|y,σ2)π(β|y,σ2) directly.
But we know π(β,σ2|y)=π(β|y,σ2)π(σ2|y)π(β,σ2|y)=π(β|y,σ2)π(σ2|y) and we already know
β|y,σ2∼Np((XTX+1νIp)−1XTy,(XTX+1νIp)−1σ2)β|y,σ2∼Np((XTX+1νIp)−1XTy,(XTX+1νIp)−1σ2)
So π(σ2|y)=∫π(σ2,β|y)dβ∝∫f(y|σ2,β)π(β|σ2)π(σ2)dβ=∫f(y|σ2,β)π(β|σ2)dβ×π(σ2)∝∫(σ2)−n2exp[−12σ2(y−Xβ)T(y−Xβ)](σ2)−p2exp(−12σ2νβTβ)dβ×π(σ2)∝∫exp[−12σ2(yTy−2βTXTy+βTXTXβ+1νβTβ)]dβ((σ2)−12(n+p)π(σ2))=∫exp[−12σ2(βT(XTX+1νIp)β−2βT(XTX+1νIp)(XTX+1νIp)−1XTy+yTy)]dβ((σ2)−12(n+p)π(σ2))=∫exp[−12σ2(β−˜β)TA(β−˜β)+yTy−˜βTA˜β]dβ((σ2)−12(n+p)π(σ2))=∫exp[−12σ2(β−˜β)TA(β−˜β)]dβ×exp[−12σ2(yTy−yTXA−1XTy)]((σ2)−12(n+p)π(σ2))=(2π)p2|σ2A−1|12exp[−12σ2(yT(In−XA−1XT)y)]((σ2)−12(n+p)π(σ2))∝(σ2)p2−12(n+p)(σ2)−a0−1exp[−1σ2(b0+12yT(In−XA−1XT)y)]=(σ2)−(12n+a0)−1exp[−1σ2(b0+12yT(In−XA−1XT)y)]=(σ2)−a∗−1exp(−b∗σ2)π(σ2|y)=∫π(σ2,β|y)dβ∝∫f(y|σ2,β)π(β|σ2)π(σ2)dβ=∫f(y|σ2,β)π(β|σ2)dβ×π(σ2)∝∫(σ2)−n2exp[−12σ2(y−Xβ)T(y−Xβ)](σ2)−p2exp(−12σ2νβTβ)dβ×π(σ2)∝∫exp[−12σ2(yTy−2βTXTy+βTXTXβ+1νβTβ)]dβ((σ2)−12(n+p)π(σ2))=∫exp[−12σ2(βT(XTX+1νIp)β−2βT(XTX+1νIp)(XTX+1νIp)−1XTy+yTy)]dβ((σ2)−12(n+p)π(σ2))=∫exp[−12σ2(β−~β)TA(β−~β)+yTy−~βTA~β]dβ((σ2)−12(n+p)π(σ2))=∫exp[−12σ2(β−~β)TA(β−~β)]dβ×exp[−12σ2(yTy−yTXA−1XTy)]((σ2)−12(n+p)π(σ2))=(2π)p2|σ2A−1|12exp[−12σ2(yT(In−XA−1XT)y)]((σ2)−12(n+p)π(σ2))∝(σ2)p2−12(n+p)(σ2)−a0−1exp[−1σ2(b0+12yT(In−XA−1XT)y)]=(σ2)−(12n+a0)−1exp[−1σ2(b0+12yT(In−XA−1XT)y)]=(σ2)−a∗−1exp(−b∗σ2) where A=(XTX+1νIp),˜β=A−1xTy,a∗=12n+a0,b∗=b0+12yT(In−XA−1XT)yA=(XTX+1νIp),~β=A−1xTy,a∗=12n+a0,b∗=b0+12yT(In−XA−1XT)y.
This is proportional to the pdf of IG(a∗,b∗)IG(a∗,b∗). Hence we have E(β|y)=∫βπ(β|y)dβ=∫β[∫π(β,σ2|y)dσ2]dβ=∫∫β[π(β|y,σ2)π(σ2|y)dσ2]dβ,E(β|y)=∫βπ(β|y)dβ=∫β[∫π(β,σ2|y)dσ2]dβ=∫∫β[π(β|y,σ2)π(σ2|y)dσ2]dβ, Similarly, Var(β|y)=∫(β−E(β|y))π(β|y)dβ=∫(β−E(β|y))[∫π(β,σ2|y)dσ2]dβ=∫∫(β−E(β|y))[π(β|y,σ2)π(σ2|y)dσ2]dβ,Var(β|y)=∫(β−E(β|y))π(β|y)dβ=∫(β−E(β|y))[∫π(β,σ2|y)dσ2]dβ=∫∫(β−E(β|y))[π(β|y,σ2)π(σ2|y)dσ2]dβ,
Plug-in Sampling Method
We can estimate mean of ββ by computing E(β|y)=∑Mm=1β(m)/ME(β|y)=∑Mm=1β(m)/M, where β(m)β(m) are generaed as follows:
- Generate σ2(m)σ2(m) from π(σ2|y)π(σ2|y), where σ2|yσ2|y is from IG(a∗,b∗)IG(a∗,b∗)
- For given σ2(m)σ2(m), generate β(m)β(m) from π(β|y,σ2(m))π(β|y,σ2(m)), where it is from normal distribution in Eq. (2)
Simulation Results
We use the same setup in the Section Importance Sampling Simulation. The final result is as follows:| E(β0)E(β0) | E(β1)E(β1) | Var(β0)Var(β0) | Var(β1)Var(β1) | |
|---|---|---|---|---|
| 100 | 1.1099 | 1.9960 | 0.0073 | 0.0107 |
| 1000 | 1.1049 | 2.0004 | 0.0079 | 0.0084 |
| 5000 | 1.1081 | 1.9966 | 0.0081 | 0.0088 |
Markov Chain Monte Carlo Simulation
Gibbs Samping
Suppose σ2σ2 is unknown, we know σ2|y,β0,β1∼IG(a∗,b∗)σ2|y,β0,β1∼IG(a∗,b∗) β|y,σ2∼Np((XTX+1νIp)−1XTy,(XTX+1νIp)−1σ2)β|y,σ2∼Np((XTX+1νIp)−1XTy,(XTX+1νIp)−1σ2) hence we can get the conditional distribution of each parameter, β0|β1,y,σ2∼N(μ0+σ0σ1ρ(β1−μ1),(1−ρ2)σ21)β0|β1,y,σ2∼N(μ0+σ0σ1ρ(β1−μ1),(1−ρ2)σ21) β1|β0,y,σ2∼N(μ1+σ1σ0ρ(β0−μ0),(1−ρ2)σ20)β1|β0,y,σ2∼N(μ1+σ1σ0ρ(β0−μ0),(1−ρ2)σ20) where σ0=[1,0]A−1σ2[1,0]Tσ0=[1,0]A−1σ2[1,0]T, σ1=[0,1]A−1σ2[0,1]Tσ1=[0,1]A−1σ2[0,1]T, μ0=A−1XTy[1,0]Tμ0=A−1XTy[1,0]T and μ1=A−1XTy[0,1]Tμ1=A−1XTy[0,1]T .
To generate β(m)β(m) from π(β|y)π(β|y), we can use a MCMC method of Gibbs sampling. The Gibbs sampling, which is one of the most popular MCMC methods, can be implemented as follows:
Set the initial value (β(0)0,β(0)1,σ2(0))(β(0)0,β(0)1,σ2(0)). Then iterate the following steps fort=0,1,2,…t=0,1,2,….
Step1: Generate σ2(t+1)σ2(t+1) from π(σ2|y,β(t)0,β(t)1)π(σ2|y,β(t)0,β(t)1).
Step2: Generate β(t+1)0β(t+1)0 from π(β0|y,β(t)1,σ2(t+1))π(β0|y,β(t)1,σ2(t+1)).
Step3: Generate β(t+1)1β(t+1)1 from π(β1|y,β(t+1)0,σ2(t+1))π(β1|y,β(t+1)0,σ2(t+1)).
We setup the total sample size is 5000, and burning period is 2000.
The final result is as follows:
| E(β0)E(β0) | E(β1)E(β1) | Var(β0)Var(β0) | Var(β1)Var(β1) | |
|---|---|---|---|---|
| 5000 | 1.1096 | 1.9955 | 0.0082 | 0.009 |
The diagnosis tool to assess the convergence of the sampler is as follows:


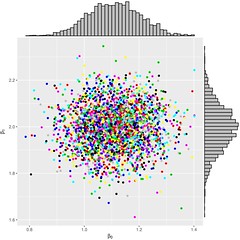
Figure 1: β0β0; β1β1; the marginals and the joint distribution
Bayesian V.S. Frequentist Case Study
Multivariable Normal Conditional Distribution
- The conditional posterior of βjβj is obtained as follows: π(βj|β−j,σ2,y)∝π(βj,β−j|σ2,y)=π(β|σ2,y)∝f(β,y|σ2)=f(y|β,σ2)π(β|σ2)∝exp(−12σ2∥y−Xβ∥2)exp(−12σ2ν∥β∥2)∝exp[−12σ2((y−X−jβ−j−xjβj)T(y−X−jβ−j−xjβj)+1νβTjβj)]∝exp[−12σ2(−yTxjβj+βT−jXT−jxjβj−βTjxTjy+βTjxTjX−jβ−j+βTjxTjxjβj+1νβTjβj)]∝exp[−12σ2(−2βTjxTjy+2βTjxTjX−jβ−j+βTjxTjxjβj+1νβTjβj)]=exp[−12σ2(βTj(xTjxj+1ν)βj−2βTjxTj(y−X−jβ−j))]=exp[−12σ2(xTjxj+1ν)(βTjβj−2βTj(xTjxj+1ν)−1xTj(y−X−jβ−j))]=exp[−12σ2(xTjxj+1ν)(βj−˜βj)2]π(βj|β−j,σ2,y)∝π(βj,β−j|σ2,y)=π(β|σ2,y)∝f(β,y|σ2)=f(y|β,σ2)π(β|σ2)∝exp(−12σ2∥y−Xβ∥2)exp(−12σ2ν∥β∥2)∝exp[−12σ2((y−X−jβ−j−xjβj)T(y−X−jβ−j−xjβj)+1νβTjβj)]∝exp[−12σ2(−yTxjβj+βT−jXT−jxjβj−βTjxTjy+βTjxTjX−jβ−j+βTjxTjxjβj+1νβTjβj)]∝exp[−12σ2(−2βTjxTjy+2βTjxTjX−jβ−j+βTjxTjxjβj+1νβTjβj)]=exp[−12σ2(βTj(xTjxj+1ν)βj−2βTjxTj(y−X−jβ−j))]=exp[−12σ2(xTjxj+1ν)(βTjβj−2βTj(xTjxj+1ν)−1xTj(y−X−jβ−j))]=exp[−12σ2(xTjxj+1ν)(βj−~βj)2] where ˜βj=(xTjxj+1ν)−1xTj(y−X−jβ−j)~βj=(xTjxj+1ν)−1xTj(y−X−jβ−j), X−jX−j is a submatrix of XX without the jthjth column, and β−jβ−j is a subvector of ββ without the jthjth element. Hence
βj|β−j,σ2,y∼N((xTjxj+1ν)−1xTj(y−X−jβ−j),σ2[xTjxj+1ν]−1)βj|β−j,σ2,y∼N((xTjxj+1ν)−1xTj(y−X−jβ−j),σ2[xTjxj+1ν]−1)
Setup
- Consider
f(y|β,σ2)=(2πσ2)−n2exp(−12σ2∥y−Xβ∥2),f(y|β,σ2)=(2πσ2)−n2exp(−12σ2∥y−Xβ∥2), π(β|σ2)=(2πσ2ν)−p2exp(−12σ2ν∥β∥2).π(β|σ2)=(2πσ2ν)−p2exp(−12σ2ν∥β∥2).
Generate 30 samples from y=1+1×x1+2×x2+0×x3+0×x4+10×x5+ϵy=1+1×x1+2×x2+0×x3+0×x4+10×x5+ϵ where ϵ∼N(0,2),xi∼N(0,1).β=(1,1,2,0,0,10)ϵ∼N(0,2),xi∼N(0,1).β=(1,1,2,0,0,10)
use frequentist way method to estimate ββ
use Bayesian MC
use Bayesian MCMC
Computation Algorithm
In frequentist way, that is OLS method, we know the estimate of ββ is ˆβ=(XTX)−1XTy^β=(XTX)−1XTy. The mean square of error is ∥ˆβ−β∥2/6∥^β−β∥2/6.
In Bayesian MC way, we assume that
σ2∼IG(a0,b0)σ2∼IG(a0,b0)
β|σ2∼N(0,σ2νIp)β|σ2∼N(0,σ2νIp)
Then we have proved that
σ2|y∼IG(a∗,b∗)σ2|y∼IG(a∗,b∗), where a∗=n2+a0,b∗=b0+12yT(In−XA−1XT)ya∗=n2+a0,b∗=b0+12yT(In−XA−1XT)y
β|y,σ2∼N(A−1XTy,A−1σ2)β|y,σ2∼N(A−1XTy,A−1σ2), where A=(XTX+1νIp)A=(XTX+1νIp)
hence β(m)β(m) are generaed as follows:Finally, we estimate ˆβ^β by computing E(β|y)=∑Mm=1β(m)/ME(β|y)=∑Mm=1β(m)/M. The mean square of error is ∥ˆβ−β∥2/6∥^β−β∥2/6.
In Bayesian MCMC way, we have proved that
- σ2|y,β∼IG(a∗,b∗), where a∗=n+p2+a0,b∗=b0+12∥y−Xβ∥2+12ν∥β∥2
- β|y,σ2∼N(A−1XTy,A−1σ2),where A=(XTX+1νIp)
- βj|β−j,σ2,y∼N((xTjxj+1ν)−1xTj(y−X−jβ−j),σ2[xTjxj+1ν]−1)
Then, we use Gibbs sampling method to get the β(m)=(β(m)0,β(m)1,…,β(m)5). Set the initial values (β(0)0,β(0)1,…,β(0)5,σ2(0)).Then iterate the following steps for t=0,1,2,… as follows:
- Generate σ2(t+1) from π(σ2|y,β(t)0,β(t)1,…,β(t)5).
- Generate β(t+1)0 from π(β0|y,β(t)−0,σ2(t+1)).
- update β(t) with β(t)=(β(t+1)0,β(t)1,…,β(t)5)
- Generate β(t+1)1 from π(β1|y,β(t)−1,σ2(t+1)).
- update β(t) with β(t)=(β(t+1)0,β(t+1)1,β(t)2,…,β(t)5) and similarly, generate β(t+1)2,…,β(t+1)5 and update β(t) after generating β(t+1)i every time,
where β(t)−i is a subvector of β without ith element.
We setup the total sample size is 5000, and burning period is 2000.
Finally, we estimate ˆβ by computing E(β|y)=∑Mm=2001β(m)/M. The mean square of error is ∥ˆβ−β∥2/6.
Simulation Results
| β0 | β1 | β2 | β3 | β4 | β5 | MSE | |
|---|---|---|---|---|---|---|---|
| Frequentist | 1.194 | 0.991 | 2.080 | -0.082 | -0.051 | 9.608 | 0.035 |
| Bayesian MC | 1.195 | 0.972 | 2.084 | -0.090 | -0.068 | 9.570 | 0.041 |
| Bayesian MCMC | 1.187 | 0.986 | 2.071 | -0.085 | -0.058 | 9.581 | 0.038 |
Model Selection
Model Selection
Consider we have k candidate models, M={M1,M2,…,Mk}. A Bayesian way to select the best model is to find the model which maximizes the model posterior probability given data, that is
ˆM=argmaxM∈Mπ(M|y) where π(M|y)=f(y|M)π(M)∑˜M∈Mf(y|˜M)π(˜M) in which, the denominator is free of M. note that the prior π(M)=1/k. Then we have π(M|y)=f(y|M)∑w∈Mf(y|˜M) This leads to ˆM=argmaxM∈Mf(y|M) where f(y|M)=∫f(y|θM,M)π(θM)dθM, where θM is the parameter under model M. We call f(y|M) as marginal likelihood under model M,f(y|θM,M) likelihood under model M and π(θM) prior under model M. Under M f(y|M)=∫f(y|θ,M)π(θ|M)dθ=∫∫f(y|β,σ2,M)π(β|σ2,M)π(σ2|M)dβdσ2=∫∫(2π)−n2(σ2)−n2exp[−12σ2(y−XβM)T(y−XβM)]×(2π)−pM2(σ2ν)−pM2exp(−12σ2νβTMβM)×ba11Γ(a1)(σ2)(−a1−1)exp(−b1σ2)dβMdσ2=ba11Γ(a1)νpM2(2π)(−n+pM2)∫∫σ2(−(n+pM+2a1+2)2)×exp[−12σ2(yTy−2βTMXTy+βTMXTXβM+1νβTMβM+2b1)]dβMdσ2 Note that yTy−2βTMXTy+βTMXTXβM+1νβTMβM+2b1=βTM(XTX+1νIpM)βM−2βTM(XTX+1νIpM)(XTX+1νIpM)−1XTy+yTy+2b1=βTMAβM−2βTMA~βM+~βMTA~βM−~βMTA~βM+yTy+2b1=(βM−~βM)TA(β−~βM)+yTy−yTXA−1XTy+2b1=(βM−~βM)TA(βM−~βM)+yT(In−XA−1XT)y+2b1 Therefore, f(y|M)=ba11Γ(a1)νpM2(2π)(−n+pM2)∫∫σ2(−(n+pM+2a1+2)2)×exp[−12σ2((βM−~βM)TA(βM−~βM)+yT(In−XA−1XT)y+2b1)]dβMdσ2=ba11Γ(a1)νpM2(2π)(−n+pM2)∫σ2(−(n+pM+2a1+2)2)×(2π)pM2|A|−12exp[−12σ2(yT(In−XA−1XT)y+2b1)]dσ2=ba11Γ(a1)νpM2(2π)(−n2)|A|−12∫σ2(−a∗M−1)exp(−b∗Mσ2)dσ2=ba11Γ(a1)νpM2(2π)(−n2)|A|−12Γ(a∗M)b∗(−a∗M)M=(2π)(−n2)Γ(a∗M)b∗(−a∗M)Mba11Γ(a1)νpM2|A|−12 where A=XTX+1νIpM,~βM=A−1XTya∗M=n+pM2+a1,b∗M=12yT(In−XA−1XT)y+b1 After Laplace approximation, f(y|M) is equivalent to BIC for large n.
MCMC
Minghui Chen (IWDE)
Goal is to compute m(y)=∫f(y|θ)π(θ)dθ
π(θ|y)=f(y|θ)π(θ)∫f(y|θ)π(θ)dθ Let g(θ) be a density function of θ
Then,
∫g(θ)dθ=1⇔∫g(θ)π(θ|y)π(θ|y)dθ=1⇔∫g(θ)π(θ|y)f(y|θ)π(θ)m(y)dθ=1⇔m(y)∫g(θ)f(y|θ)π(θ)π(θ|y)dθ=1⇔m(y)=[∫g(θ)f(y|θ)π(θ)π(θ|y)dθ]−1=E[g(θ)f(y|θ)π(θ)|y]−1≈[M∑m=1g(θ(m))f(y|θ(m))π(θ(m))/M]−1 where θ(m) is from π(θ|y).
There maxmimum of m(y) is equivalent to maximize E[g(θ)f(y|θ)π(θ)|y]−1
Now let’s use Minghui Chen’s method to maximize the f(y|Mi).
f(y|Mi)=∫f(y|θ,Mi)π(θ|Mi)dθ, where θ=(β0,β1,σ2) π(θ|y,Mi)=f(y|θ,Mi)π(θ|Mi)∫f(y|θ,Mi)π(θ|Mi)dθ=f(y|θ,Mi)π(θ|Mi)f(y|Mi) Let g(θ) be a density function of θ
Then
∫g(θ)dθ=1⇔∫g(θ)π(θ|y,Mi)π(θ|y,Mi)dθ=1⇔∫g(θ)π(θ|y,Mi)f(y|θ,Mi)π(θ|Mi)f(y|Mi)dθ=1⇔f(y|Mi)∫g(θ)f(y|θ,Mi)π(θ|Mi)π(θ|y,Mi)dθ=1⇔f(y|Mi)=[∫g(θ)f(y|θ,Mi)π(θ|Mi)π(θ|y,Mi)dθ]−1=E[g(θ)f(y|θ,Mi)π(θ|Mi)|y,Mi]−1≈[N∑n=1g(θ(n))f(y|θ(n),Mi)π(θ(n)|Mi)/N]−1 where θ(n) is from π(θ|y,Mi),i=1,2,…,k.
The issue here is a choice of g(θ) to minimize MC error.
Recall π(θ|y)∝f(y|θ)π(θ)≈g(θ) Let g(θ) be a normal density such that Eg(θ)= posterior mean ≈∑Mm=1θ(m)/M=ˉθ, Vg(θ)= posterior variance matrix ≈∑Mm=1(θ(m)−ˉθ)(θ(m)−ˉθ)T/M, where θ(m) is a MCMC sample from π(θ|y).
For given model M, using MCMC, generate θ(k)M from π(θM|y,M), then compute ˉθM and VM.
Define g(θ|M)=N(θ;ˉθM,VM).
Compute
f(y|M)≈[K∑k=1g(θ(k)|M)f(y|θ(k),M)π(θ(k)|M)/K]−1
Bayesian Simulation Case Study
Setup
- Consider
f(y|β,σ2)=(2πσ2)−n2exp(−12σ2∥y−Xβ∥2), π(β|σ2)=(2πσ2ν)−p2exp(−12σ2ν∥β∥2).
Generate 30 samples from y=1+1×x1+2×x2+0×x3+0×x4+10×x5+ϵ where ϵ∼N(0,2),xi∼N(0,1).β=(1,1,2,0,0,10)
Therefore, the true model, say Mture, is y=1+1×x1+2×x2+10×x5+ϵ
- use Bayesian MC method to find true model
- use frequentist way method to find true model
Our candidate models are as follows: M1:y=β0+ϵ; M2:y=β0+β1X1+ϵ; … M32:y=1+1×x1+2×x2+0×x3+0×x4+10×x5+ϵ
- For bayesian way, compute
f(y|Mk) for k=1,2,…,32 and select the best model ˆM, such that f(y|ˆM)≥f(y|Mk) If ˆM=Mtrue, then assign Zi=1, otherwise Zi=0.
Repeat the above procedure for R=500 times, and compute ∑Ri=1ZiR≃P( selecting true model with Bayes method ).
- For Frequentist way, similarly, build linear model for 32 candidate models, compute AIC and BIC. If the smallest AIC or BIC is ture model, then Wi=1, otherwise Wi=0. Repeat it 500 times and compute ∑Ri=1WiR.
Consider the sample sizes are 20, 50, 100 and 300 so that we can see how sample size impacts the probability of selecting the true model.
- For MCMC, sampling with MCMC method, then compute Eq. (4) for 32 candidates models, select the best model ˆM, such that f(y|ˆM)≥f(y|Mk) If ˆM=Mtrue, then assign Zi=1, otherwise Zi=0
Repeat the above procedure for R=500 times, and compute ∑Ri=1ZiR≃P( selecting true model with MCMC method )
Result
| MC | AIC | BIC | MCMC | Error | |
|---|---|---|---|---|---|
| 20 | 0.502 | 0.560 | 0.656 | 0.508 | 59.401 |
| 50 | 0.894 | 0.668 | 0.882 | 0.880 | 78.224 |
| 100 | 0.998 | 0.676 | 0.916 | 0.926 | 93.176 |
| 300 | 0.998 | 0.712 | 0.978 | NA | NA |